Match! 粉末衍射軟體

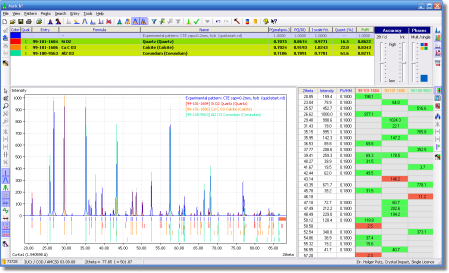
Match! is an easy-to-use software for phase identification from powder diffraction data, which has become a daily task in material scientists work. Match! compares the powder diffraction pattern of your sample to a database containing reference patterns in order to identify the phases which are present. Single as well as multiple phases can be identified based on both peak data and raw (profile) data.
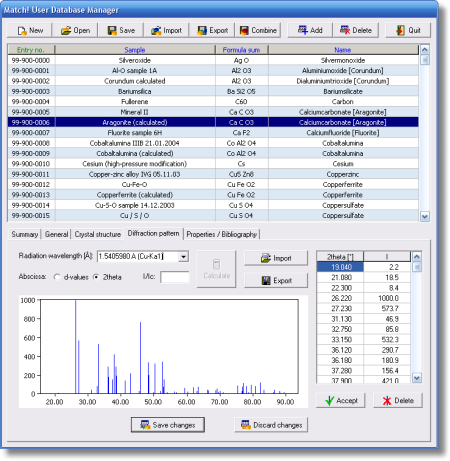
As reference database, you can apply the included free-of-charge IUCr/COD/AMCSD database and/or ICSD/Retrieve (if you have a valid licence), use any ICDD PDF product, and/or create a user database based on your own diffraction patterns. The user database patterns can be edited manually, imported from peak files, calculated from crystal structure data (e.g. CIF files), or imported from your colleague's user database.
Match! Function List
- Fast single and multiple phase identification from powder diffraction data
- Use free-of-charge reference patterns calculated from the IUCr/ COD/AMCSD (incl. I/Ic), any ICSD/Retrieve version (released 1993-2002; valid licence required), any ICDD PDF database and/or your own diffraction data (or patterns calculated from crystal structure data (e.g. CIF files)) in phase identification
- Flexible handling of reference databases (incl. user databases); you can easily switch between different reference databases without the necessity to perform a new database indexation
- Create reference databases for neutron diffraction e.g. from cif-files
- Comfortable user database manager for easy maintenance of user data (add/import/edit/delete/sort entries)
- Powerful CIF- and ICSD/Retrieve import, incl. calculation of powder pattern, I/Ic and density
- Atomic coordinates available e.g. in the ICSD, the ICDD PDF-4+ or free-of-charge reference data are displayed in the data sheets and included in the CIF exports (e.g. for Rietveld analysis)
- View crystal structures in Diamond with just a few clicks
- Fully integrated handling of your own diffraction data with PDF data (search-match, retrieval, data viewing)
- Automatic residual searching with respect to identified phases
- Automatic raw data processing: α2-stripping, background subtraction, peak search, profile fitting, error correction
- Automatic optimization of peak searching sensitivity
- Fitting of all (or selected) peak parameters to exp. profile data
- Comfortable manual editing of peaks (add/shift/delete/fit) using mouse or keyboard
- Semi-quantitative analysis (Reference Intensity Ratio method)
- Straight-forward usage of additional knowledge (composition, PDF subfiles, crystallographic data, color, density etc.)
- Integrated database retrieval system and viewer for PDF and user datasheets
- Multiple step undo/redo
- Support for new PDF entry numbers (9 digits)
- User-configurable automatic operation
- Full parameter control with instant rearrangement of the results list
- Automatic d-value shifting during search-match process (optionally)
- Intensity contribution to figure-of-merit can be reduced for preferred-orientation cases
- Comfortable graphical and tabular comparison of peak data and candidate patterns
- Mouse-controlled pattern graphics zooming and tracking
- User-configurable HTML-reports
- Online update (automatic or manual)
- Supported diffraction data file formats (automatic detection):
- ASCII profile (start, step, intensities or 2 columns)
- Bruker/Siemens raw data (old and new) (*.raw)
- Bruker/Siemens DIFFRAC AT peak data (*.dif)
- DBWS (*.rfl, *.dat)
- ENDEAVOUR peak list (2 columns: 2theta/d intensity; *.dif)
- Inel raw data (*.dat)
- Ital Structures raw data (*.esg)
- Jade/MDI/SCINTAG raw data (*.mdi)
- JEOL ASCII Export raw data (*.txt)
- PANalytical XRDML Scan raw data (*.xrdml)
- PANalytical/Philips peak data (*.udi)
- PANalytical/Philips raw data (*.rd, *.udf)
- Rigaku raw data (*.raw)
- SCINTAG raw data (*.raw, *.rd)
- Shimadzu raw data (*.raw)
- Siemens (*.uxd)
- Sietronics XRD scan data (*.cpi)
- Stoe raw data (*.raw)
- Stoe peak data (*.pks)
System requirements
- Personal Computer with Microsoft Windows 98, ME, NT 4.0, 2000, XP or Vista operating system
- Microsoft Internet Explorer 5.01 or higher
- 128 megabytes of RAM (256 megabytes recommended)
- Hard disk with minimum 600 MB of free disk space
- Graphics resolution of 1024 x 768 pixels with 32,768 colors ("High Color")
- DVD drive